Release date:2020-10-23
JACI
[IF:6.771]
The international sinonasal microbiome study: A multicentre, multinational characterization of sinonasal bacterial ecologyDOI: 10.1111/all.14276
Abstract:
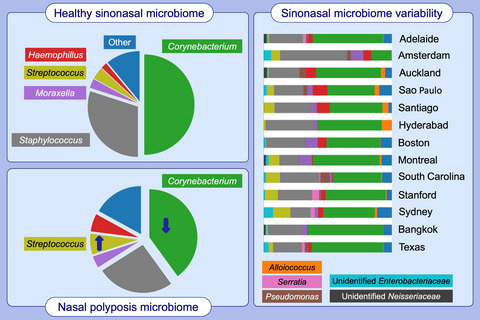
The sinonasal microbiome remains poorly defined, with our current knowledge based on a few cohort studies whose findings are inconsistent. Furthermore, the variability of the sinus microbiome across geographical divides remains unexplored. We characterize the sinonasal microbiome and its geographical variations in both health and disease using 16S rRNA gene sequencing of 410 individuals from across the world. Although the sinus microbial ecology is highly variable between individuals, we identify a core microbiome comprised of Corynebacterium, Staphylococcus, Streptococcus, Haemophilus and Moraxella species in both healthy and chronic rhinosinusitis (CRS) cohorts. Corynebacterium (mean relative abundance = 44.02%) and Staphylococcus (mean relative abundance = 27.34%) appear particularly dominant in the majority of patients sampled. Amongst patients suffering from CRS with nasal polyps, a statistically significant reduction in relative abundance of Corynebacterium (40.29% vs 50.43%; P = .02) was identified. Despite some measured differences in microbiome composition and diversity between some of the participating centres in our cohort, these differences would not alter the general pattern of core organisms described. Nevertheless, atypical or unusual organisms reported in short‐read amplicon sequencing studies and that are not part of the core microbiome should be interpreted with caution. The delineation of the sinonasal microbiome and standardized methodology described within our study will enable further characterization and translational application of the sinus microbiota.
First Author:
Sathish Paramasivan
Correspondence:
Alkis J. Psaltis, Department of Otolaryngology, Head and Neck Surgery, The Queen Elizabeth Hospital, 28 Woodville Road, Woodville South, SA 5011, Australia.
All Authors:
Sathish Paramasivan, Ahmed Bassiouni, Arron Shiffer, Matthew R. Dillon, Emily K. Cope, Clare Cooksley, Mahnaz Ramezanpour, Sophia Moraitis, Mohammad Javed Ali, Benjamin Bleier, Claudio Callejas, Marjolein E. Cornet, Richard G. Douglas, Daniel Dutra, Christos Georgalas, Richard J. Harvey, Peter H. Hwang, Amber U. Luong, Rodney J. Schlosser, Pongsakorn Tantilipikorn, Marc A. Tewfik, Sarah Vreugde, Peter-John Wormald, J. Gregory Caporaso, Alkis J. Psaltis
2020-09-03 Article
 hth官方网页版中国有限公司
hth官方网页版中国有限公司
